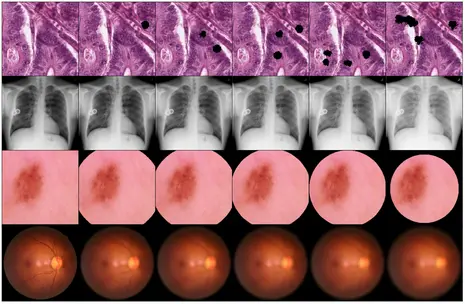
ADSMI Workshop Paper: "MedMNIST-C: Comprehensive benchmark and improved classifier robustness by simulating realistic image corruptions"
The integration of neural networks into clinical practice is constrained by challenges related to domain generalization and robustness. To address these issues, MedMNIST-C, a comprehensive benchmark dataset based on the MedMNIST+ collection, has been introduced. MedMNIST-C covers 12 datasets and 9 imaging modalities, simulating task- and modality-specific image corruptions of varying severity to evaluate algorithm robustness against real-world artifacts and distribution shifts. Quantitative evidence shows that easy-to-use artificial corruptions enable high-performance, lightweight data augmentation, improving model robustness. Unlike traditional, generic augmentation strategies, this approach leverages domain knowledge and demonstrates significantly higher robustness.
For more details, you can access the paper here and the associated code repository here.
Main Conference Paper: "Self-supervised Vision Transformer are Scalable Generative Models for Domain Generalization"
Despite notable advancements, the integration of deep learning (DL) techniques into clinically relevant applications, especially in digital histopathology, has been hindered by challenges related to robust generalization across various imaging domains and features. Traditional mitigation strategies in this area, such as data augmentation and stain color normalization, have proven insufficient, necessitating the exploration of alternative methods. To address this, a novel generative method for domain generalization in histopathology images is proposed. This method employs a generative, self-supervised Vision Transformer to dynamically extract features from image patches and seamlessly integrate them into the original images, thereby creating novel synthetic images with diverse attributes. By enriching the dataset with such synthesized images, the aim is to enhance its holistic nature, facilitating improved generalization of DL models to unseen domains. Extensive experiments conducted on two distinct histopathology datasets demonstrate the effectiveness of this approach, significantly outperforming the state of the art, both on the Camelyon17-wilds challenge dataset (+2%) and on a second epithelium-stroma dataset (+26%). Additionally, the method's ability to easily scale with the increasing availability of unlabeled data samples and more complex, higher-parametric architectures is highlighted.
For more details, you can access the paper here and the associated code repository here.
About MICCAI and ADSMI
The MICCAI conference is recognized as a premier annual event focusing on the latest developments in medical image computing, computer-assisted intervention, and related fields. It attracts top-tier research from around the world, fostering collaboration and advancements in medical imaging AI.
The MICCAI Workshop on Advancing Data Solutions in Medical Imaging AI (ADSMI) provides a platform for discussion and innovation at the intersection of medical imaging, AI, and data science. This workshop addresses challenges of data scarcity, quality, and interoperability in medical imaging AI, bringing together efforts from Data Augmentation, Labeling, and Imperfections, Big Task Small Data, and Medical Image Learning with Limited and Noisy Data.
Stay tuned for exciting insights and updates directly from MICCAI 2024 in October!